
Tuberculosis (TB) is one of the top ten causes of death worldwide. In 2017, 10 million people around the world fell ill with TB and 1.3 million died. The genome of the bacterium that causes TB holds a special toxin-antitoxin system with spectacular action: once the toxin is activated, all bacterial cells die, stopping the disease. An international research team co-led by
the Wilmanns group at EMBL in Hamburg investigated this promising feature for therapeutic targets. They now share the first high-resolution details of the system in Molecular Cell.
Mycobacterium tuberculosis is the bacterium that causes TB in humans. Its genome holds 80 so-called toxin-antitoxin (TA) systems: sets of closely linked genes that encode both a toxic protein and an antitoxin: a toxin-neutralising antidote.
When the bacteria are growing normally, toxin activity is blocked by the antitoxin’s presence. But under stress conditions such as lack of nutrients, dedicated enzymes rapidly degrade the antitoxin molecules. This activates the toxin proteins in the cell and slows down the growth of the bacteria, allowing them to survive the stressful environment.

Novel NAD+-degrading toxin triggers cell death of tuberculosis-causing bacteria. The illustration shows sections of Mycobacterium tuberculosis bacteria. The Pac-Man symbolizes the toxin, which in the absence of its antitoxin counterpart, ‘eats up’ the NAD+, causing the cell to die. In the bacterium in the back, the Pac-Man’s mouth is blocked by the antitoxin, preventing the degradation of NAD+ and allowing the cell to grow normally. Illustration by
Beata Edyta Mierzwa.
One particular TA system has a more drastic effect: in the absence of the antitoxin, the toxin kills the bacteria. As this system holds potential for therapeutic targets, researchers from EMBL Hamburg, the IPBS at the CNRS/Université de Toulouse, and the Crick Institute in London joined forces to study this TA system in more detail.
“Our goal was to see the TA system’s structure, so we could try to understand and even manipulate it. It was as if we were working blindly before”, says Annabel Parret, EMBL staff scientist in the Wilmanns group, who led the project.
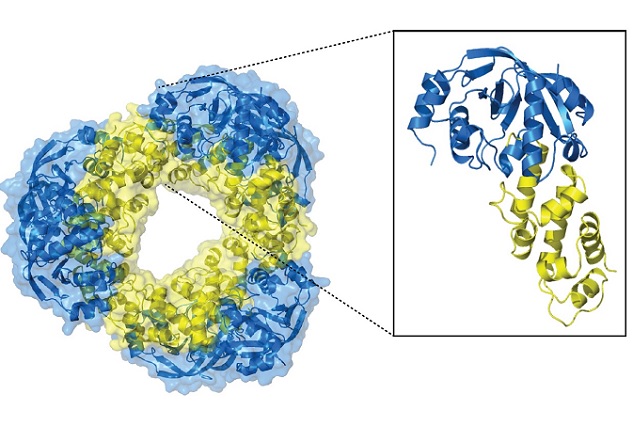
The high-resolution structure of the toxin-antitoxin system. CREDIT: EMBL Hamburg
The high-resolution structure – solved within eight months by first author Diana Freire – revealed a large and compact system with a double-doughnut shape. “It looks like a diamond, and it is very stable,” says EMBL group leader Matthias Wilmanns. The structure resembles the toxins of cholera and diphtheria: diseases that caused epidemics with hundreds of thousands of people dying even within the past 100 years.
Knowledge of the structure gave important guidance for further studying the system’s biochemistry – a challenging part of the project. By using an interdisciplinary approach, the team were able to discover the details of the TA system’s mode of action. When the toxin dissociates from its antidote, it becomes activated and starts to degrade essential cellular metabolites called NAD+ molecules. This “suicide” activity ultimately leads to the death of all bacterial cells. Why the bacteria have such a suicide system is puzzling, but there is no doubt it has the potential to be exploited as a drug target.
“Our collaborators in Toulouse were already able to extend the lifetime of mice infected with TB by activating the toxin in a controlled way,” says Parret. “If we find molecules that can disrupt the TA system – and thus trigger cell death – in TB patients, that would be the perfect drug.”

The team will now screen thousands of small molecules to see if they have this capability. However, the structure of the TA system is so stable that it will be a big challenge to find an entry point where they can go in to break it. Wilmanns: “But if we succeed, this could be a new approach for treating TB and other infectious diseases.”
This project was a collaboration between researchers from the Wilmanns group at EMBL Hamburg and the Neyrolles group at the IPBS, CNRS/Université de Toulouse, and involved the Carvalho group at the Francis Crick Institute in London. The research team has started a partnership to work towards a potential TB drug, through EMBL’s technology transfer arm EMBLEM.
Source article:
Freire, D.M., Gutierrez, C., et al. An NAD+ phosphorylase t
oxin triggers Mycobacterium tuberculosis cell death. Molecular Cell, published online 18 February 2019.

Posted by Dr. Tim Sandle,
Pharmaceutical Microbiology










